Research
Synthetic gene regulatory networks for spatiotemporal pattern formation
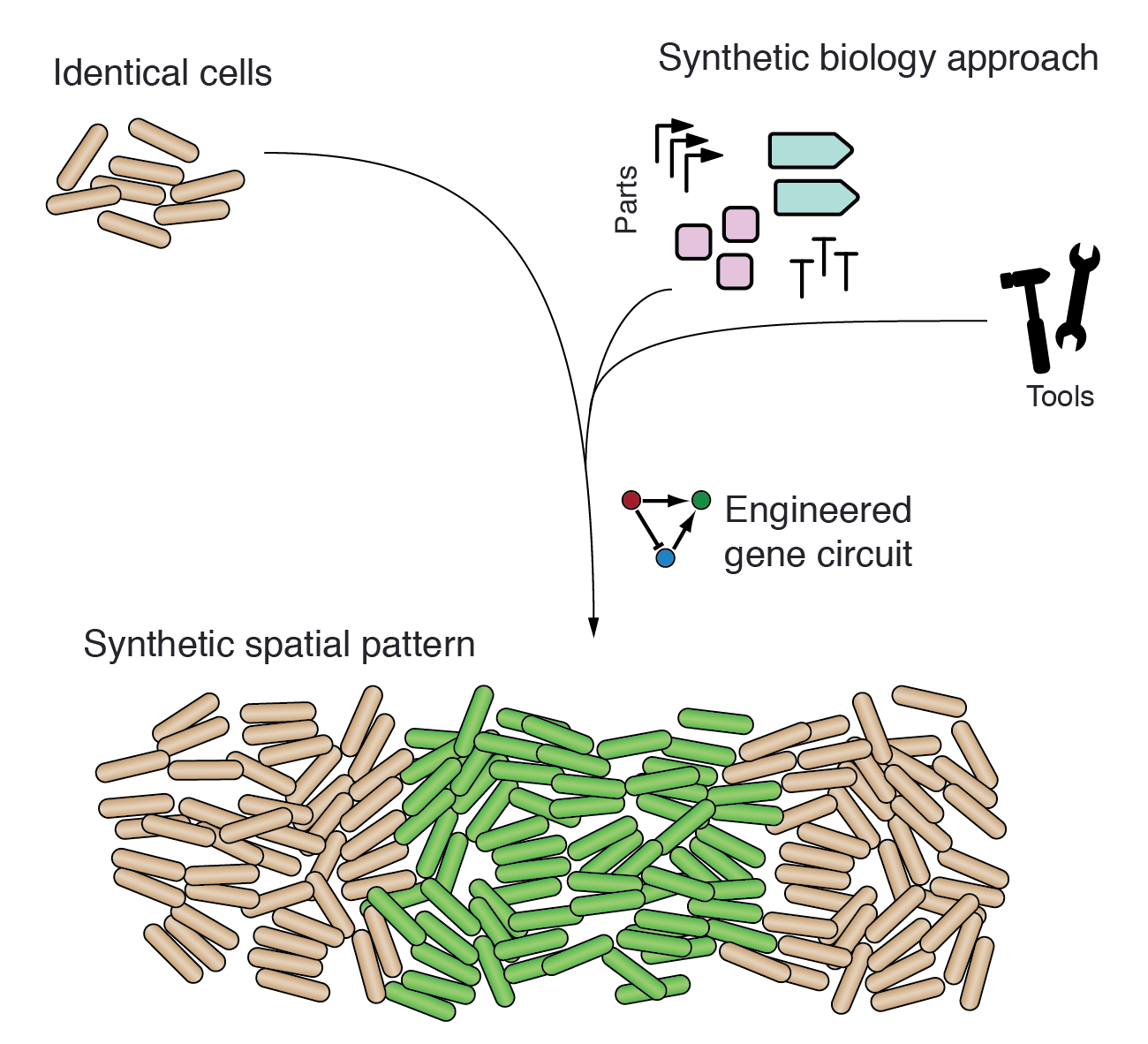
Patterns are omnipresent in biology, including patterns in microbial communities, gene expression patterns during development and skin or fur patterns of animals. The molecular details of pattern formation are complicated, but are thought to be governed by general principles such as the use of morphogen gradients to provide positional information. We build, study and model synthetic gene regulatory networks to improve our understanding of such general principles. Moreover, we aim to engineer patterns of interest in bacterial populations.
Relevant publications:
Pignon, E., Holló, G.Steiner, T., van Vliet, S., Schaerli, Y.(2025) Uptake and leakage rates differentially shape community arrangement and composition of microbial consortia, The ISME Journal, :wraf122
Park, J. H., Holló, G., Schaerli, Y. (2024) From resonance to chaos by modulating spatiotemporal patterns through a synthetic optogenetic oscillator, Nat. Commun., 15:7284
Santos-Moreno, J., Tasiudi, E., Kusumawardhani, H., Stelling, J., Schaerli, Y. (2023) Robustness and innovation in synthetic genotype networks, Nat. Commun., 14:2454
Barbier, I., Kusumawardhani, H., Schaerli, Y. (2022) Engineering synthetic spatial patterns in microbial populations and communities, Curr. Opin. Microbiol., 67:102149
Barbier, I., Perez-Carrasco, R., Schaerli, Y. (2020) Controlling spatiotemporal pattern formation in a concentration gradient with a synthetic toggle switch, Molecular Systems Biology, 16:e9361
Santos-Moreno, J., Tasiudi, E., Stelling, J., Schaerli, Y. (2020) Multistable and dynamic CRISPRi-based synthetic circuits, Nat. Commun., 11:2746
Santos-Moreno, J., Schaerli, Y. (2018) Using Synthetic Biology to Engineer Spatial Patterns, Advanced Biosystems, doi: 10.1093/nar/gkv1310
Schaerli, Y., Munteanu, A.Gili, M.Cotterell, J.Sharpe, J.Isalan, M. (2014) A unified design space of synthetic stripe-forming networks, Nat. Commun., 5:4905
Novel tools and approaches for synthetic biology
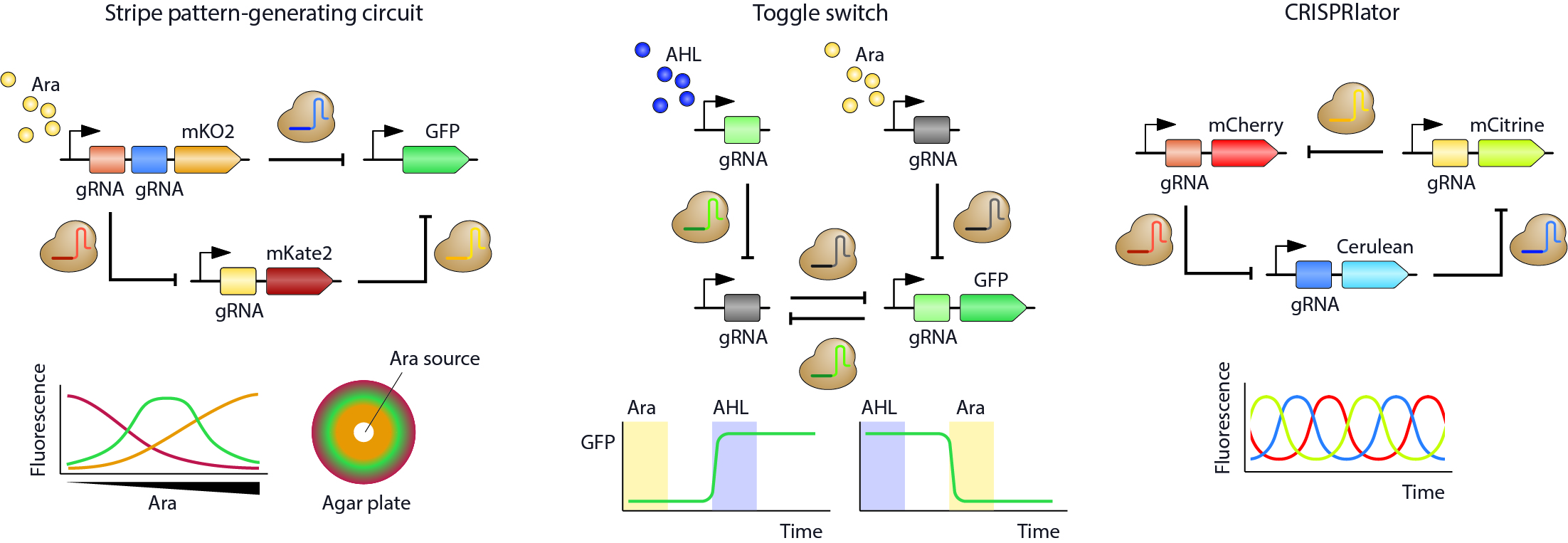
We develop novel tools and approaches for synthetic biology. We were the first to demonstrate that CRISPR interference (CRISPRi) can be used to build dynamic and multistable synthetic gene circuits, such as a bistable toggle switch and an oscillator (“CRISPRlator”). These exciting results show that it is feasible to apply CRISPRi widely in synthetic circuits and paves the way for engineering more complex synthetic networks. For example, we have now built 3-node networks with up to 6 interactions and also combined CRISPRi with CRISPR activation (CRISPRa). The use of CRISPR-based circuits promises to bypass many of the limitations that synthetic biologists are currently facing with protein-based circuits. This will allow us to build complex, modular and versatile synthetic circuits.
Relevant publications:
Kusumawardhani, H., Zoppi, F., Avendaño, R., Schaerli, Y. (2025) Engineering Intercellular Communication using M13 Phagemid and CRISPR-based Gene Regulation for Multicellular Computing in Escherichia coli, Nat. Commun., 16:3569
Santos-Moreno, J., Tasiudi, E., Kusumawardhani, H., Stelling, J., Schaerli, Y. (2023) Robustness and innovation in synthetic genotype networks, Nat. Commun., 14:2454
Barbier, I., Kusumawardhani, H., Chauhan, L., Harlapur, P. V., Jolly, M. K., Schaerli, Y. (2023) Synthetic gene circuits combining CRISPR interference and CRISPR activation in E. coli: importance of equal guide RNA binding affinities to avoid context-dependent effects, ACS Synth. Biol., 12, 10, 3064–3071
Santos-Moreno, J., Schaerli, Y. (2020) CRISPR-based gene expression control for synthetic gene circuits, Biochem Soc Trans, 48:1979 48:-1993
Duarte, J. M., Barbier, I., Schaerli, Y. (2017) Bacterial microcolonies in gel beads for high-throughput screening of libraries in synthetic biology, ACS Synth. Biol., 6:1988-1995
Patterned engineered living materials
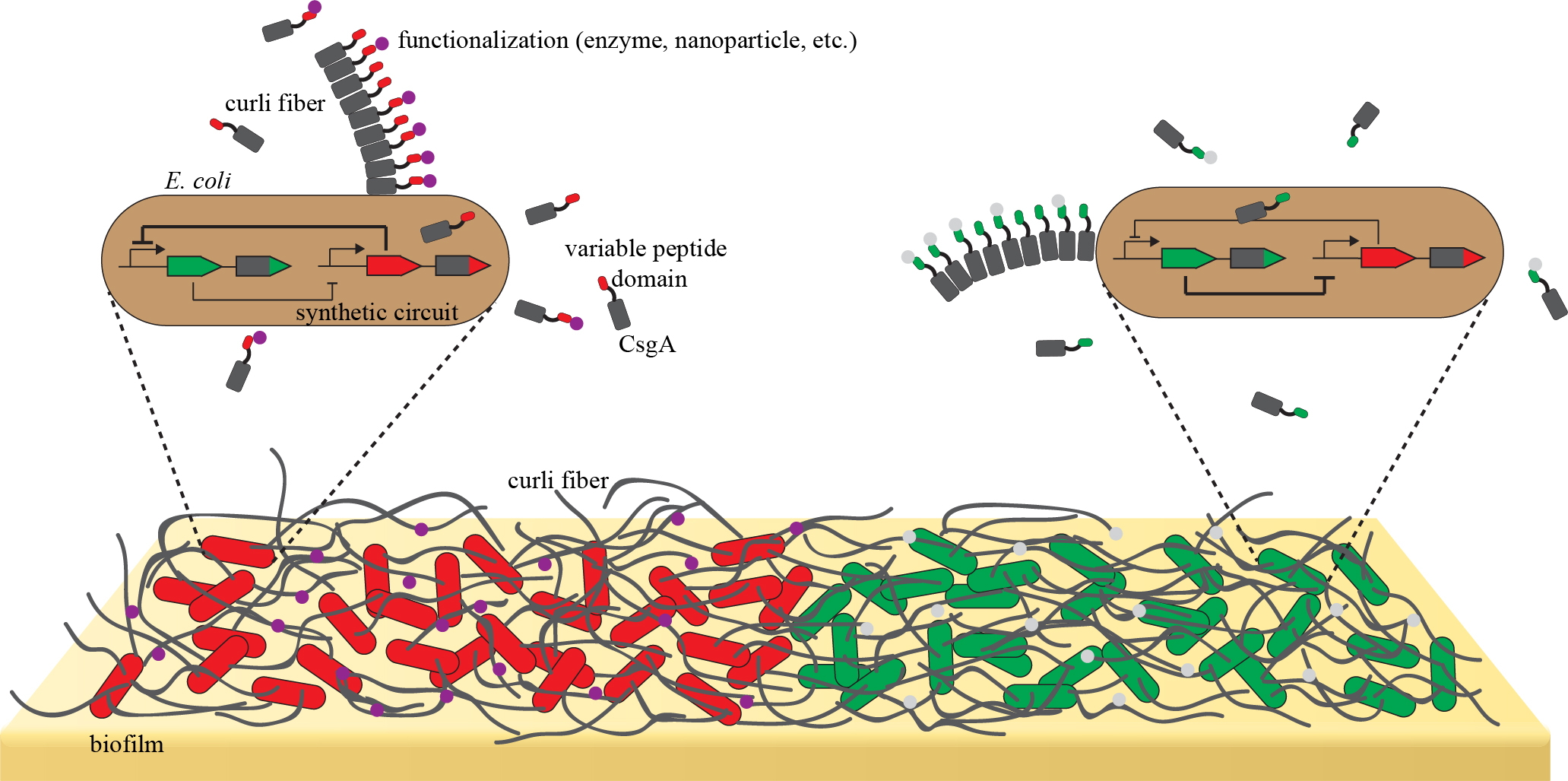
Our ability to build synthetic circuits controlling spatiotemporal gene expression is of interest for applications. We started working on one of them, namely engineered living materials. Indeed, biological systems possess properties that are highly desirable for the next generation of materials: they emerge through self-organisation controlled by a genetic program, they can self-repair and adapt to environmental cues, and they are driven by sustainable energy sources and biodegradable. Thus, engineering cells to produce user-defined living materials with these properties has the potential to contribute to a more sustainable future. Bacterial biofilms are an ideal platform for such “engineered living materials”. In biofilms, bacteria are embedded in an extracellular matrix, which dictates the physicochemical properties of the biofilm. Particularly the composition of the proteinaceous fraction is highly programmable and enables functionalisation e.g. with enzymes or nanoparticles. However, in order to realise the full potential of these materials, we need to be able to control their spatiotemporal organisation. We aim to establish a synthetic biology platform to control E. coli biofilms that self-organise in space and time to produce user-defined patterns. We are currently focusing on biofilms displaying CsgA curli fibres fused to peptide domains enabling the production of functionalised materials.
Bacteria-host interactions
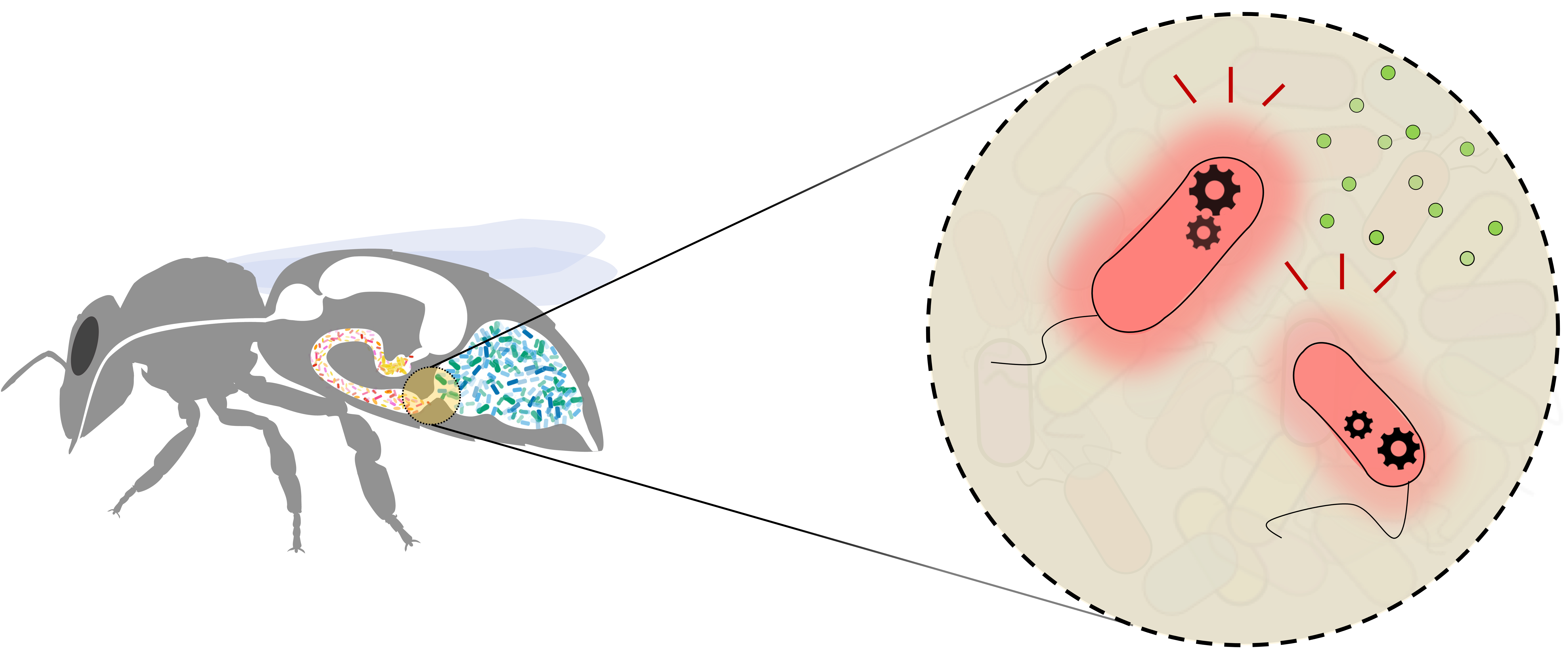
As part of the NCCR microbiomes and in collaboration with the group of Prof. Engel at the University of Lausanne, we are engineering synthetic circuits in Snodgrassela alvi a member of the honeybee gut microbiota. These tools will enable rapid building of engineered bacteria to answer fundamental questions in host-gut microbiota research.
Relevant publications:
Chhun, A., Moriano-Gutierrez, S., Zoppi, F., Cabirol, A., Engel, P., Schaerli, Y. (2023) Engineering a symbiont as a biosensor for the honey bee gut environment, bioRxiv, doi: https://doi.org/10.1101/2023.02.02.526826
Evolution of gene regulatory networks
What constrains the evolution of gene regulatory networks? What makes a gene regulatory network robust to mutations and/or evolvable? How does the regulatory mechanism of a network influence its evolution? How can gene regulatory networks change extensively, while maintaining overall circuit output? How do mutations in gene regulatory networks interact to produce novel phenotypes? What happens after a gene has been duplicated?
We have several projects where we address such questions with synthetic gene regulatory networks in E. coli.
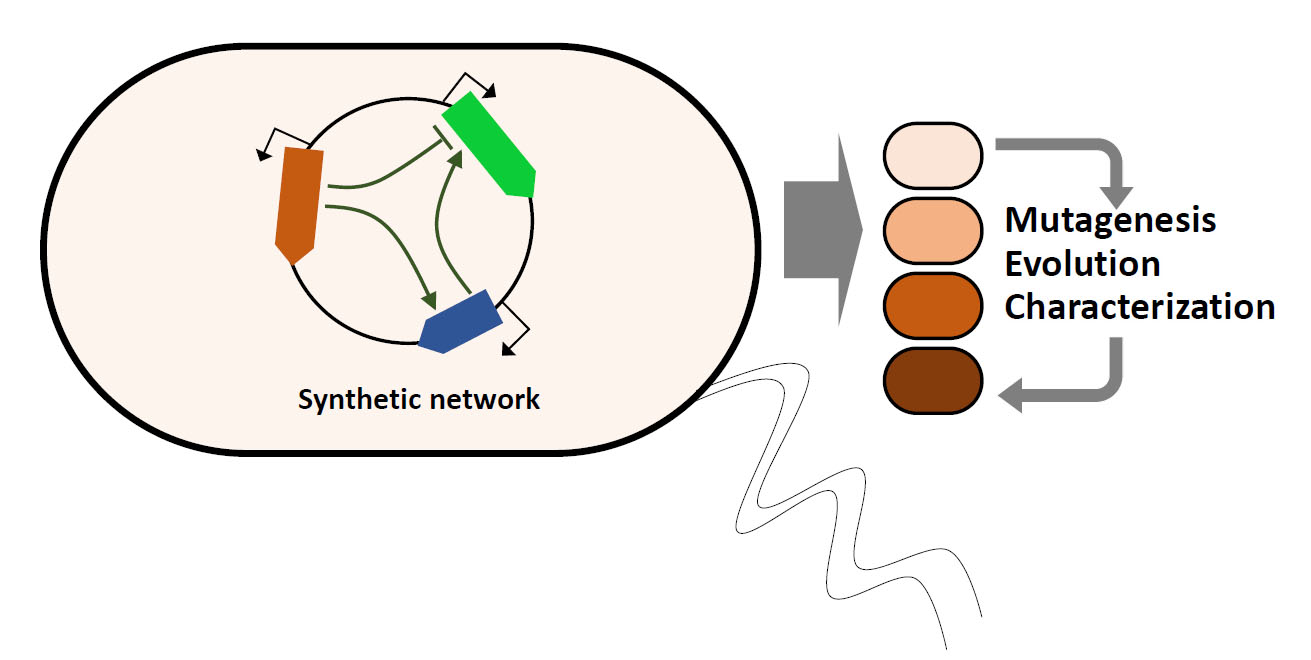
Relevant publications:
Mihajlovic, L., Iyengar, B. R., Baier, F., Barbier, I., Iwaszkiewicz, J., Zoete, V., Wagner, A., Schaerli, Y. (2025) A direct experimental test of Ohno’s hypothesis, eLife, 13:RP97216 v
Santos-Moreno, J., Tasiudi, E., Kusumawardhani, H., Stelling, J., Schaerli, Y. (2023) Robustness and innovation in synthetic genotype networks, Nat. Commun., 14:2454
Baier, F., Gauye, F., Perez-Carrasco, R., Payne, J.L., Schaerli, Y. (2023) Environment-dependent epistasis increases phenotypic diversity in gene regulatory networks, Sci. Adv., 9:eadf1773
Schaerli, Y., Jimenez, A., Duarte, J. M., Mihajlovic, L., Renggli, J., Isalan, M., Sharpe, J., Wagner, A. (2018) Synthetic circuits reveal how mechanisms of gene regulatory networks constrain evolution, Molecular Systems Biology, 14:e8102
Duarte, J. M., Barbier, I., Schaerli, Y. (2017) Bacterial microcolonies in gel beads for high-throughput screening of libraries in synthetic biology, ACS Synthetic biology, doi: 10.1021/acssynbio.7b00111
Funding
The lab currently receives funding from UNIL, SNSF and the NCCR microbiomes .

